Real-time PCR, also known as qRT-PCR, is crucial in many labs. It’s a powerful tool used to amplify and measure DNA.
But, like any complex technique, qRT-PCR can have its challenges. Troubleshooting issues can be tricky and time-consuming. Whether you are facing unexpected results or technical glitches, understanding common problems is essential. This guide will help you identify and solve common qRT-PCR issues.
With clear steps and practical tips, you’ll be back on track quickly. Let’s dive in and make your qRT-PCR experiments more reliable and efficient.
Credit: www.sigmaaldrich.com
Introduction To Qrt-pcr
Quantitative Reverse Transcription PCR, or qRT-PCR, is a powerful tool in molecular biology. Scientists use it to measure RNA levels in samples. This technique helps understand gene expression patterns. It is crucial for various research applications.
Basics Of Qrt-pcr
qRT-PCR combines reverse transcription and polymerase chain reaction. First, reverse transcription converts RNA into complementary DNA (cDNA). Then, PCR amplifies this cDNA. The process uses specific primers and probes. These components ensure accurate detection and quantification.
Fluorescent dyes or probes monitor amplification in real-time. This allows scientists to measure the amount of target RNA. The more RNA present, the more fluorescence detected. This correlation helps determine gene expression levels.
Importance In Research
qRT-PCR is essential in various fields of research. It aids in studying gene expression related to diseases. Scientists can compare gene expression between healthy and diseased tissues. This comparison helps identify potential biomarkers.
qRT-PCR is also valuable in virology. Researchers use it to detect and quantify viral RNA. This is crucial for diagnosing infections and monitoring viral load. Accurate data helps in developing treatments and vaccines.
Environmental studies benefit from qRT-PCR too. Scientists analyze microbial communities in different environments. Understanding these communities helps in conservation and environmental management.
Common Qrt-pcr Issues
Quantitative Real-Time PCR (qRT-PCR) is a powerful technique used in molecular biology to amplify and quantify DNA. However, like all techniques, it comes with its fair share of challenges. Understanding these common qRT-PCR issues can significantly improve your results and save you a lot of time and resources. Let’s dive into some of the most common problems you might encounter.
Amplification Failures
Amplification failures can be a real headache. Imagine running your qRT-PCR, only to find that none of your samples amplified! This can be caused by several factors:
- Primer Design: Poorly designed primers can lead to non-specific binding or no binding at all. Ensure that your primers are specific to your target sequence and have a melting temperature (Tm) within the optimal range.
- Template Quality: The quality of your DNA or RNA template is crucial. Contaminants like proteins, phenol, or ethanol can inhibit the PCR reaction. Make sure your template is pure and properly quantified.
- Reagent Quality: Always use high-quality reagents. Expired or improperly stored reagents can lead to failed amplifications.
- Thermocycler Issues: Ensure your thermocycler is calibrated correctly. A malfunctioning thermocycler can cause inconsistent results.
Low Sensitivity
Low sensitivity in qRT-PCR means that you might not be detecting low-abundance targets effectively. Here are some common causes and solutions:
- Suboptimal Primer Concentrations: If the concentration of your primers is too low, it can lead to inefficient amplification. Conversely, too high concentrations can lead to non-specific amplification.
- Inadequate Template Amount: Ensure you are using an adequate amount of template. Too little template can lead to low sensitivity. However, avoid using too much template as it can also cause inhibition.
- Inhibitors in the Reaction: Inhibitors can come from various sources, including the sample itself or the reagents used. It might be helpful to purify your template further or to use an inhibitor-resistant polymerase.
- Master Mix Components: The components of your master mix, including the polymerase, dNTPs, and buffers, need to be optimized for your specific reaction. Sometimes, tweaking these components can significantly improve sensitivity.
Troubleshooting qRT-PCR can sometimes feel like detective work, but understanding these common issues and how to address them can make the process much smoother. Have you faced any of these issues in your experiments? How did you solve them? Share your experiences in the comments below!
Sample Preparation Tips
Sample preparation is crucial for successful qRT-PCR experiments. Proper sample preparation ensures accurate and reliable results. Let’s dive into some essential tips.
Rna Extraction Best Practices
Start with fresh, high-quality samples. Use a reliable extraction kit. Follow the manufacturer’s instructions closely. Ensure the integrity of RNA. Use RNase-free tubes and tips. Maintain a clean work area. Avoid repeated freeze-thaw cycles of RNA. Store RNA at -80°C until use. Verify RNA quality with a spectrophotometer. Aim for a 260/280 ratio of 1.8-2.0.
Avoiding Contamination
Contamination can ruin your qRT-PCR results. Work in a clean environment. Use dedicated pipettes for RNA work. Wear gloves at all times. Change gloves frequently. Use aerosol-resistant tips. Treat surfaces with RNase inhibitors. Include negative controls in your experiments. Regularly clean equipment and workspaces. Avoid talking while pipetting. Be mindful of your work environment to prevent contamination.
Primer Design Strategies
When it comes to qRT-PCR troubleshooting, primer design is like the cornerstone of a sturdy building. It’s essential. But, how do you make sure your primers are up to the task? Let’s dive into some tried-and-true strategies that can save you from pulling your hair out in frustration.
Choosing The Right Primers
Imagine you’re baking a cake. You wouldn’t use salt instead of sugar, right? The same goes for choosing primers. The right primers are crucial for specific amplification. Here are a few tips:
- Specificity: Ensure that primers are specific to your target sequence. This reduces the chance of amplifying the wrong sequence.
- Length: Aim for primers that are 18-25 nucleotides long. This length is a sweet spot for specificity and efficiency.
- GC Content: A GC content of 40-60% helps primers bind more stably to the DNA. Too high or too low can cause issues.
Remember that poorly chosen primers can lead to non-specific bands or no amplification at all. It’s like trying to pick a lock with a spoon – frustrating and ineffective!
Optimizing Primer Concentration
You’ve got your primers, now what? Optimizing their concentration is the next step. Think of it like seasoning a dish. Too little, and it’s bland. Too much, and it’s overpowering.
- Start with a standard concentration: Typically, 0.1-0.5 µM is a good starting point.
- Tweak as needed: If amplification is weak, increase the concentration slightly. If you see non-specific bands, decrease it.
- Consistency is key: Always use the same concentration for your experiments to maintain reproducibility.
To put it simply, finding the right balance can make a world of difference. It’s like Goldilocks finding the porridge that’s just right.
By following these primer design strategies, you’ll be well on your way to qRT-PCR success. Remember, a little attention to detail now can save you a lot of headache later on.
Reaction Optimization
Reaction optimization is crucial for successful qRT-PCR experiments. Fine-tuning various parameters can significantly improve the efficiency and accuracy of your results. In this section, we will focus on adjusting annealing temperatures and optimizing MgCl2 concentration. These steps can help you achieve optimal reaction conditions for your specific assay.
Adjusting Annealing Temperatures
Annealing temperature affects primer binding specificity. Too high, and primers may not bind. Too low, and non-specific binding can occur. Start with a gradient PCR to determine the optimal temperature. Use a range of temperatures around the calculated melting temperature (Tm) of your primers. Observe the results and choose the temperature that provides the best specificity and yield.
Optimizing Mgcl2 Concentration
MgCl2 is essential for DNA polymerase activity. It affects enzyme function and primer binding. Too little MgCl2 can lead to weak or no amplification. Too much can cause non-specific amplification. Begin with the manufacturer’s recommended concentration. Then, test different concentrations to find the optimal one. Typically, 1.5 to 3.0 mM is a good starting range. Adjust as needed based on your results.
Credit: www.idtdna.com
Data Analysis Techniques
When it comes to qRT-PCR, data analysis can sometimes feel like navigating a maze. How do you make sense of those numbers and ensure your results are accurate? In this section, we’ll walk through some essential techniques for analyzing your qRT-PCR data. From interpreting Ct values to normalizing gene expression, you’ll find practical tips to make your data analysis process smoother and more reliable.
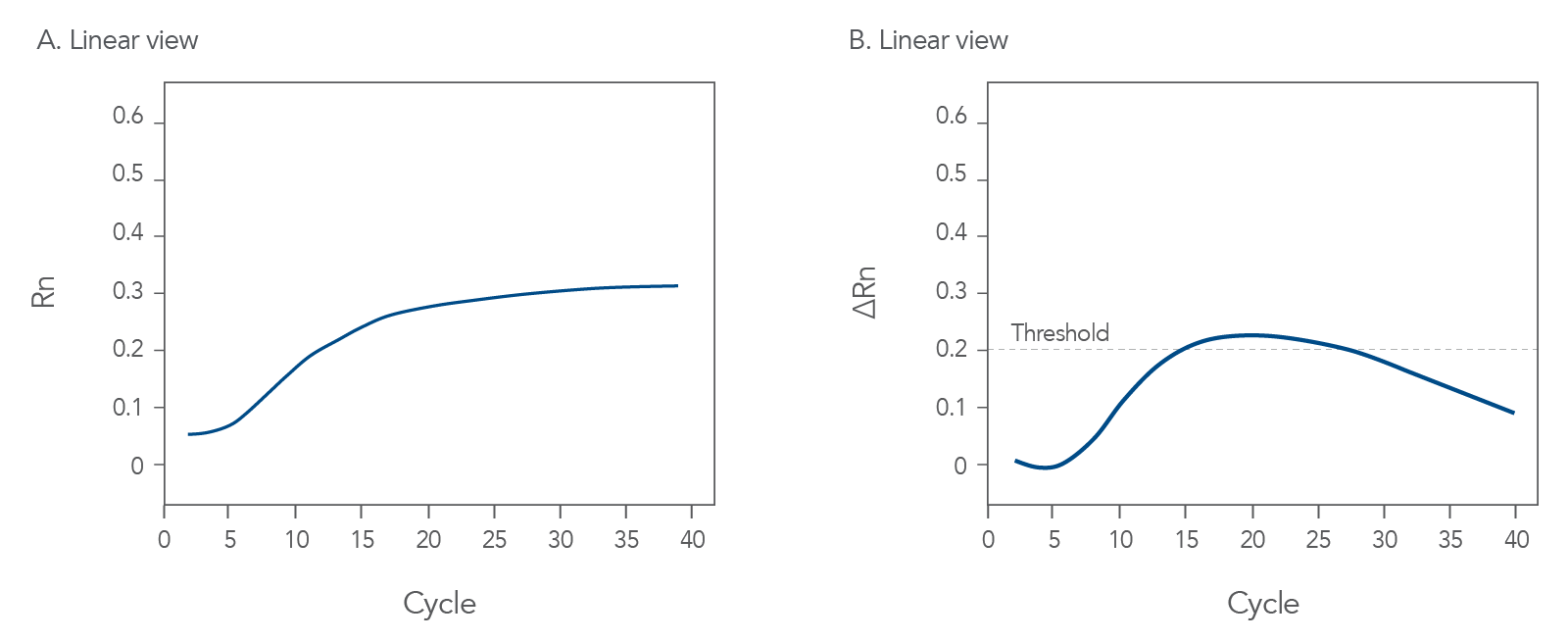
Interpreting Ct Values
First things first, let’s talk about Ct values. Ct, or threshold cycle, is the point at which your sample’s fluorescence rises above the background noise. It’s a critical number because it tells you how much of your target gene is present.
Here’s a simple way to think about it: the lower the Ct value, the more of your target gene you have. Conversely, a high Ct value means there’s less of your target gene. But what if your Ct values are all over the place? Here’s a quick guide:
- Consistent Ct values: Great! Your experiment is likely running smoothly.
- High variability: Uh-oh. This could indicate pipetting errors or inconsistent sample preparation.
- Very high Ct values: Your target gene might be in low abundance, or there could be issues with primer efficiency.
Keep an eye out for these patterns. They can provide valuable clues about what’s happening in your experiment.
Normalizing Gene Expression
Now, let’s dive into normalizing gene expression. This step is crucial because it helps you account for any variability that might occur during your qRT-PCR experiment. Think of it like calibrating a scale before weighing something—normalization ensures that your measurements are accurate.
To normalize gene expression, you’ll typically use one or more housekeeping genes. These are genes that are stably expressed across different samples and experimental conditions. Common housekeeping genes include GAPDH, ACTB, and 18S rRNA. Here’s how you can normalize your data:
- Choose an appropriate housekeeping gene for your experiment.
- Calculate the ΔCt (Delta Ct) value:
ΔCt = Ct(target gene) – Ct(housekeeping gene) - Use the ΔCt value to compare gene expression levels across your samples.
By normalizing to a housekeeping gene, you can reduce the impact of any technical variations and get a clearer picture of your target gene’s expression.
So there you have it—two essential techniques for analyzing qRT-PCR data. With these tips, you’ll be well on your way to making sense of your results and troubleshooting any issues that arise. Happy experimenting!
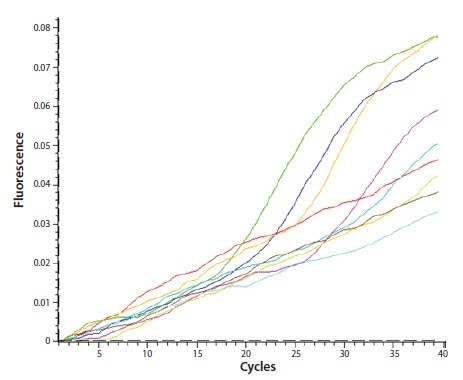
Troubleshooting Amplification Curves
Troubleshooting amplification curves in qRT-PCR can be challenging. These curves provide vital information about your reactions. Problems with these curves can lead to inaccurate results. Addressing these issues ensures accurate data and successful experiments.
Identifying Non-specific Peaks
Non-specific peaks indicate possible contamination or primer-dimer formations. Check the melting curve analysis to identify these peaks. Specific peaks should appear at a single temperature. Multiple peaks may suggest non-specific products.
Optimize your primer design to reduce non-specific amplification. Increase the annealing temperature or adjust the primer concentration. Clean your workspace and use fresh reagents. This reduces the risk of contamination.
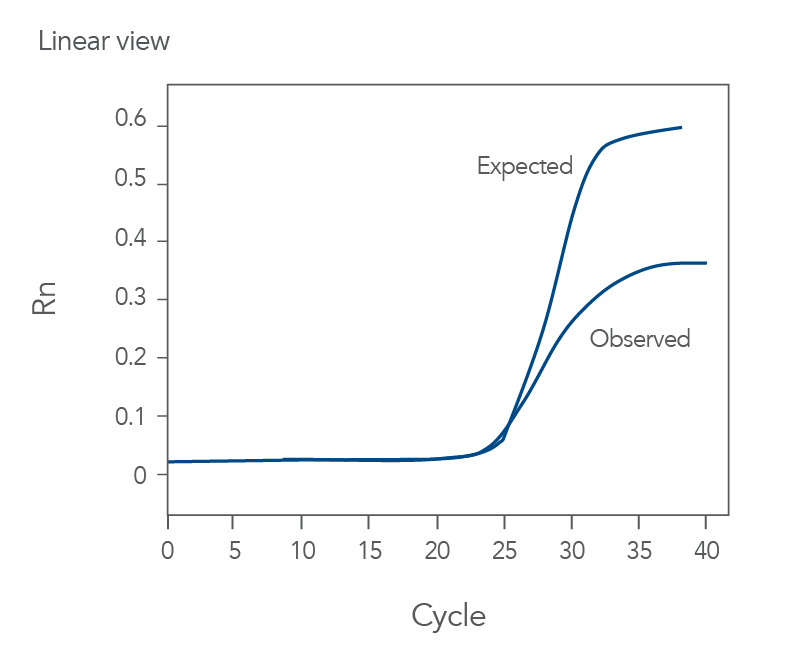
Addressing Plateau Effects
Plateau effects occur when amplification efficiency declines in later cycles. This can affect the accuracy of your quantification. Ensure your reagents are fresh and properly stored. Check the reaction components for any issues.
Review the cycling conditions and adjust if necessary. Increase the enzyme concentration or optimize the reaction mix. This can help maintain amplification efficiency. Ensure your template quality is high. Degraded templates can contribute to plateau effects.
Instrument And Software Maintenance
Maintaining your qRT-PCR instrument and software is crucial for accurate results. Neglecting maintenance can lead to errors, which can be both frustrating and time-consuming. Let’s dive into some essential maintenance tips that will keep your instrument running smoothly and your software up to date.
Regular Calibration
Think of calibration as the regular check-up your qRT-PCR machine needs. Just like your car needs a tune-up, your instrument needs calibration. It ensures that the machine’s measurements are accurate and reliable.
Why is calibration important? Without proper calibration, the results you get could be off. This could lead to incorrect conclusions and wasted resources. Regular calibration helps prevent this.
- Schedule calibration every 6 months.
- Use standard reference materials to check accuracy.
- Document each calibration session for future reference.
By following these steps, you can trust your machine’s results and avoid any unexpected surprises.
Software Updates
Software updates might seem like a hassle, but they are vital for your qRT-PCR system’s performance. These updates can fix bugs, improve features, and enhance security.
Why should you update the software? Imagine using an outdated map to find a new place; you might get lost. Similarly, outdated software can lead to errors and inefficiencies.
- Check for updates regularly.
- Read the update notes to understand improvements.
- Backup your data before updating.
Keeping your software updated ensures that your qRT-PCR system operates at its best, providing accurate and reliable results.
In conclusion, maintaining your qRT-PCR instrument and software is like taking care of a beloved pet; it requires regular attention and care. By calibrating your instrument and updating your software, you’ll ensure your experiments run smoothly and your results are trustworthy. Happy troubleshooting!
Advanced Troubleshooting Tips
When it comes to Qrt-PCR, achieving flawless results often requires more than just following a protocol. Sometimes, even the most experienced scientists encounter unforeseen issues. This is where advanced troubleshooting tips come in handy. In this section, we will explore techniques to overcome common obstacles and ensure your experiments run smoothly. Ready to dive in? Let’s get started!
Dealing With Inhibitors
Inhibitors can be the bane of any Qrt-PCR experiment. They interfere with the reaction, leading to poor or inconsistent results. Here are some tips to tackle them:
- Sample Purity: Ensure your samples are free of contaminants. Use purification kits designed specifically for RNA or DNA extraction.
- Optimize Lysis Conditions: Adjust the lysis buffer and incubation times to minimize the presence of inhibitors.
- Use Inhibitor-Resistant Enzymes: Some polymerases are more resistant to inhibitors. Consider switching to these if you frequently encounter issues.
- Dilute Samples: Sometimes, simply diluting your sample can reduce the concentration of inhibitors to a manageable level.
Enhancing Reaction Efficiency
Boosting the efficiency of your Qrt-PCR reactions can significantly improve your results. Here are some practical strategies:
- Optimize Primer Design: Ensure your primers are specific and efficient. Use online tools to help design the best primers for your target sequence.
- Master Mix Adjustments: Fine-tune the components of your master mix. Sometimes, slight adjustments in the concentrations of MgCl2, dNTPs, or enzymes can make a big difference.
- Cycling Conditions: Optimize the cycling conditions, including the annealing temperature and extension time. This can enhance the amplification efficiency.
- Template Quality: Use high-quality, intact RNA or DNA. Degraded templates can lead to inefficient reactions.
Remember, troubleshooting Qrt-PCR is often about making small tweaks and observing their effects. By systematically addressing potential issues, you can enhance the reliability and accuracy of your experiments.
Have you encountered any unique challenges with Qrt-PCR? Share your experiences and tips in the comments below!
Credit: www.idtdna.com
Conclusion And Best Practices
qPCR troubleshooting is essential for accurate results. Understanding common issues and their fixes can save time and resources. This section covers key points and final recommendations. Follow these best practices for consistent and reliable qPCR data.
Summary Of Key Points
Ensure the use of high-quality reagents. Always prepare fresh master mixes. Validate primers and probes for specificity. Check for contamination regularly. Optimize annealing temperatures and cycle numbers. Maintain a clean workspace.
Final Recommendations
Consistency is key. Use the same reagents and protocols. Document every step for traceability. Regularly calibrate your qPCR machine. Train all users thoroughly. Keep a detailed log of all runs. This helps identify patterns and issues quickly. Double-check results with replicates. They can confirm data accuracy.
By following these practices, you improve the reliability of your qPCR results. Troubleshooting becomes easier and less frequent. Accurate data leads to better research outcomes.
Frequently Asked Questions
What Are Three Common Reasons For The Failure Of A Pcr Reaction?
Three common reasons for PCR failure are poor primer design, contaminated reagents, and incorrect thermal cycling conditions.
Why Isn’t My Qpcr Working?
Your qPCR might not be working due to incorrect reagent preparation, poor sample quality, or pipetting errors. Verify primer specificity and optimize reaction conditions. Ensure proper thermal cycler settings and troubleshoot any equipment issues.
What Are The Sources Of Error In Qpcr?
Sources of error in qPCR include pipetting mistakes, sample contamination, primer-dimer formation, and inefficient reverse transcription. Using poor quality reagents or degraded RNA can also cause issues.
What Are The Possible Errors In Pcr Amplification?
Possible errors in PCR amplification include primer-dimer formation, non-specific binding, contamination, and primer mismatches. These errors affect accuracy.
What Causes No Amplification In Qrt-pcr?
No amplification can be due to degraded RNA, inhibitors, or incorrect primer design.
Conclusion
Troubleshooting qRT-PCR can seem daunting, but it’s manageable with patience. Identify potential issues methodically. Check reagents, equipment, and protocols closely. Small adjustments often yield big improvements. Keep detailed records of each step. This helps in pinpointing problems. Stay consistent with your procedures.
Seek advice from experienced colleagues if stuck. Remember, practice makes perfect. Happy troubleshooting!
