QPCR troubleshooting involves identifying issues like primer-dimer formation and optimizing reaction conditions. Ensure accurate pipetting and proper reagent storage.
QPCR, or quantitative polymerase chain reaction, is a powerful technique for measuring DNA or RNA levels. Researchers often face challenges that can affect the accuracy of results. Common issues include primer-dimer formation, inadequate annealing temperatures, and inconsistent pipetting. Ensuring proper reagent storage and using validated primers can mitigate many problems.
Regularly calibrating equipment and maintaining a clean workspace are also crucial. By identifying and addressing these issues, you can achieve reliable and reproducible QPCR results. This guide will help you troubleshoot effectively and enhance the quality of your experiments.
Introduction To Qpcr
qPCR, also known as quantitative Polymerase Chain Reaction, is a powerful lab technique. It helps scientists amplify and quantify DNA sequences. This method is crucial in various research fields, from genetics to virology. Understanding the basics and importance of qPCR can help troubleshoot common problems.
Basics Of Qpcr
qPCR involves several key steps. First, DNA is extracted from the sample. Then, the DNA is mixed with specific reagents. These reagents include primers, nucleotides, and a DNA polymerase enzyme. The mixture is placed in a qPCR machine. The machine cycles through different temperatures. Each cycle doubles the amount of DNA.
There are two main types of qPCR: SYBR Green and TaqMan. SYBR Green binds to any double-stranded DNA. TaqMan uses a probe specific to the target DNA sequence. Both methods have their advantages. SYBR Green is simpler and cheaper. TaqMan is more specific and accurate.
Importance In Research
qPCR is a vital tool in research. It allows scientists to study gene expression. This helps in understanding disease mechanisms. qPCR is also used in diagnostics. It can detect viruses like COVID-19. The method provides rapid and accurate results.
Researchers use qPCR in various fields:
- Genetics: Studying gene mutations
- Oncology: Monitoring cancer markers
- Virology: Detecting viral infections
- Pharmacology: Evaluating drug effects on genes
qPCR’s versatility makes it indispensable in modern science.
| Field | Application |
|---|---|
| Genetics | Gene mutations |
| Oncology | Cancer markers |
| Virology | Viral infections |
| Pharmacology | Drug effects |
Common Qpcr Errors
qPCR is a sensitive technique. Small errors can lead to big problems. Knowing common errors helps prevent them. Below, we discuss two critical issues: inaccurate quantification and primer-dimer formation.
Inaccurate Quantification
Inaccurate quantification can ruin your results. Several factors cause this issue. Some common causes include:
- Poor Pipetting: Pipetting errors affect sample volumes. Use calibrated pipettes.
- Inconsistent Standards: Standard curves must be accurate. Ensure standards are well-prepared.
- Inhibitors: Presence of inhibitors can affect amplification. Purify samples thoroughly.
Ensure all reagents are fresh. Store them as per guidelines. Avoid freeze-thaw cycles.
Primer-dimer Formation
Primer-dimer formation is another common issue. Primers can bind to each other. This non-specific binding affects your results. To minimize primer-dimers:
- Design Primers Carefully: Use software tools for primer design.
- Optimize Annealing Temperature: Higher temperatures reduce non-specific bindings.
- Use Hot-Start Taq Polymerase: It activates only at high temperatures. This helps reduce primer-dimers.
Always run a no-template control (NTC). This helps identify primer-dimer issues.
Sample Quality Issues
Effective qPCR results depend heavily on the quality of your samples. Compromised sample quality can lead to inaccurate or inconclusive results. This section explores common sample quality issues that can affect qPCR outcomes.
Rna Integrity
Ensuring RNA integrity is essential for reliable qPCR results. Degraded RNA can lead to a loss of signal and biased data. Use high-quality extraction methods to maintain RNA integrity.
Check RNA quality using an Agilent Bioanalyzer or similar instrument. Look for clear and distinct rRNA peaks. Peaks indicate intact RNA. Smears suggest degradation.
| Indicator | Quality |
|---|---|
| Distinct rRNA peaks | Intact RNA |
| Smears | Degraded RNA |
Dna Contamination
DNA contamination can interfere with qPCR results. Contaminated RNA samples may produce misleading data. Use DNase treatment to remove DNA contamination from RNA samples.
- Perform a no-reverse-transcription control (No-RT control) to detect DNA contamination.
- Use RNA-specific primers to avoid amplifying DNA.
- Validate RNA purity with spectrophotometry. Check A260/A280 ratios.
An A260/A280 ratio of 1.8-2.0 indicates pure RNA. Lower ratios suggest contamination.

Credit: sg.idtdna.com
Reagent Problems
Reagent problems are a common issue in qPCR troubleshooting. Reagents include the master mix, enzymes, and buffers. Each component is crucial for the success of your qPCR experiments. Understanding reagent problems can help you achieve accurate and reliable results.
Master Mix Composition
The master mix composition plays a key role in qPCR. If the master mix is not prepared correctly, the results can be inconsistent.
- Ensure all components are fresh.
- Check the concentration of each reagent.
- Use nuclease-free water.
Sometimes, the buffer or dNTPs in the master mix can degrade. Always store these reagents at the recommended temperature. This prevents degradation and maintains their effectiveness.
Enzyme Activity
The enzyme activity is essential for successful qPCR. If the enzyme is inactive, the reaction will fail.
- Check the expiration date of the enzyme.
- Store the enzyme at -20°C to maintain activity.
- Use the recommended amount for your reaction.
Inactive enzymes can result from improper storage. Always handle enzymes with care. Avoid repeated freeze-thaw cycles to preserve their activity.
| Reagent | Common Issues | Solutions |
|---|---|---|
| Master Mix | Incorrect concentrations | Verify and adjust concentrations |
| Enzyme | Inactive enzyme | Check storage conditions |
Technical Mistakes
Technical mistakes in qPCR can lead to inaccurate results. These errors can stem from various sources. Identifying and correcting them is crucial for accurate qPCR data.
Pipetting Errors
Pipetting errors are common in qPCR. They can significantly affect the accuracy of your results. Ensuring proper technique is key.
- Use calibrated pipettes for every experiment.
- Avoid air bubbles in the pipette tips.
- Consistently pipette at the same angle.
- Use the correct pipette tip size.
Inconsistent pipetting can lead to variable sample volumes. This inconsistency can cause errors in your qPCR data. Always double-check your pipetting technique.
Thermal Cycler Calibration
The thermal cycler is the heart of the qPCR process. Proper calibration is essential for accurate results.
| Calibration Aspect | Importance |
|---|---|
| Temperature Accuracy | Ensures correct annealing and extension. |
| Uniformity | All wells should have the same temperature. |
Regular calibration of your thermal cycler is a must. Skipping this step can lead to uneven temperature distribution. This can cause significant errors in your qPCR results.
Always follow the manufacturer’s guidelines for calibration. This ensures your thermal cycler works efficiently.
Credit: www.sigmaaldrich.com
Data Analysis Challenges
Data analysis in qPCR can be tricky. It involves several critical steps. Each step impacts the accuracy of your results. Let’s dive into some common data analysis challenges.
Baseline Setting
Baseline setting is crucial in qPCR. It helps you determine the background noise. Correct baseline setting ensures accurate quantification.
Improper baseline settings can skew your data. Always check the baseline during the initial cycles. This helps in eliminating non-specific signals.
| Cycle Range | Description |
|---|---|
| 3-15 | Typical range for baseline |
| 15-40 | Amplification phase |
Threshold Determination
The threshold determines the point of detection. Setting the threshold correctly is vital for accurate results.
Threshold should be set above the background noise. This ensures only real signals are detected.
- Set the threshold in the exponential phase.
- Avoid setting it in the baseline or plateau phase.
Incorrect threshold settings can lead to false positives or negatives. Always validate your threshold settings with a control sample.
Here are some quick tips for better threshold determination:
- Use automated threshold settings as a starting point.
- Manually adjust if needed.
- Validate with multiple replicates.
Optimization Strategies
Optimization strategies are essential for successful qPCR experiments. By fine-tuning various parameters, you can achieve more accurate and reliable results. This section will explore effective optimization strategies focusing on primer design and reaction conditions.
Primer Design
Effective primer design is critical for qPCR success. Here are key tips:
- Specificity: Ensure primers bind only to your target sequence.
- Length: Primers should be 18-25 nucleotides long.
- GC Content: Maintain a GC content of 40-60%.
- Melting Temperature (Tm): Aim for a Tm of 58-60°C.
- Secondary Structures: Avoid hairpins, dimers, and repeats.
Use tools like Primer3 or NCBI Primer-BLAST for design. Validate primers using in-silico PCR tools to predict performance.
Reaction Conditions
Optimizing reaction conditions enhances qPCR efficiency. Consider the following factors:
| Factor | Recommendation |
|---|---|
| Annealing Temperature | Optimize between 55-65°C for best results. |
| Mg2+ Concentration | Test concentrations from 1.5 to 3.0 mM. |
| Template DNA | Use 1-10 ng of template DNA per reaction. |
| Cycle Number | Typically, 35-40 cycles are sufficient. |
Perform a gradient PCR to find the optimal annealing temperature. Adjust Mg2+ concentration if amplification is weak. Ensure template DNA quality by using high-purity samples.
By applying these optimization strategies, you can improve your qPCR results. Happy troubleshooting!
Case Studies
Exploring real-life case studies can help identify common qPCR troubleshooting issues. This section delves into specific examples and the lessons learned from them.
Real-life Examples
One researcher faced issues with inconsistent amplification curves. Their problem was traced to a pipetting error. They resolved this by using a calibrated pipette and following a strict protocol.
Another lab had contamination problems. Their negative controls showed unexpected signals. They identified the source as an unsterilized work area. They implemented stringent cleaning procedures and eliminated the issue.
| Issue | Cause | Solution |
|---|---|---|
| Inconsistent Amplification Curves | Pipetting Error | Use Calibrated Pipette |
| Contamination | Unsterilized Work Area | Implement Cleaning Procedures |
Lessons Learned
Consistency in pipetting is crucial for accurate qPCR results. Always use a calibrated pipette to avoid errors. Follow a strict protocol to ensure consistency.
Maintaining a clean work environment is essential. Regularly clean and sterilize your work area to prevent contamination.
- Use calibrated equipment
- Follow strict protocols
- Maintain cleanliness
These lessons highlight the importance of attention to detail and consistent practices in qPCR experiments.
Best Practices
Ensuring accurate and reliable qPCR results requires adherence to best practices. Implementing these practices helps avoid common pitfalls and enhances the reproducibility of your experiments.
Quality Control
Quality control is crucial for qPCR. Always use high-quality reagents and consumables. Check the expiration dates of all materials before use. Prepare a master mix to minimize pipetting errors.
- Use nuclease-free water.
- Test for contamination regularly.
- Ensure consistent sample storage conditions.
Run negative controls to detect contamination. Include a no-template control (NTC) in every run. This helps identify contamination in reagents or pipetting errors.
Regular Maintenance
Maintaining your qPCR instrument is key to accurate results. Regular calibration is necessary to ensure precise temperature control. Check the optical system for dust and debris.
- Clean the sample block regularly.
- Verify the calibration of the thermal cycler.
- Inspect and clean the optical system.
Keep a log of all maintenance activities. This helps track performance issues over time.
| Maintenance Task | Frequency |
|---|---|
| Calibration | Monthly |
| Optical System Cleaning | Weekly |
| Sample Block Cleaning | After Each Run |
By following these best practices, you will improve the reliability and accuracy of your qPCR experiments.
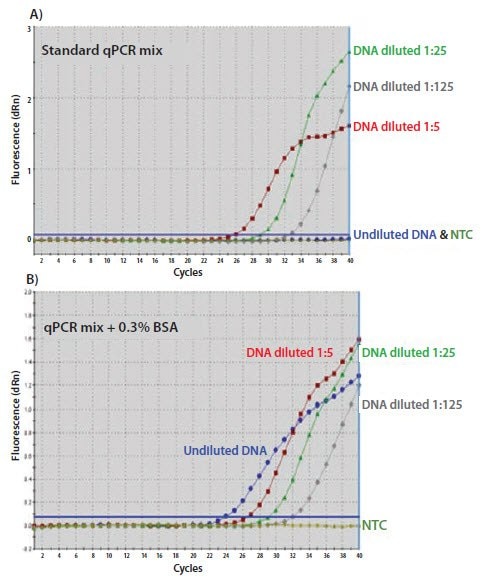
Credit: www.sigmaaldrich.com
Frequently Asked Questions
What Causes Low Qpcr Efficiency?
Low qPCR efficiency can be caused by poor primer design, degraded reagents, or suboptimal reaction conditions.
How To Fix Primer-dimer Issues?
To fix primer-dimer issues, redesign primers, optimize annealing temperature, or use a hot-start polymerase.
Why Is My Qpcr Showing No Amplification?
No amplification may result from incorrect reagent concentrations, faulty primers, or poor sample quality.
How To Improve Qpcr Reproducibility?
Improve qPCR reproducibility by using consistent sample preparation, precise pipetting, and validated reagents.
What Affects Qpcr Melting Curve Analysis?
Melting curve analysis can be affected by primer-dimers, non-specific products, or incorrect annealing temperatures.
Conclusion
Mastering qPCR troubleshooting enhances your research efficiency. Always monitor reagents, equipment, and protocols. Stay consistent with best practices. Regularly updating your knowledge ensures accurate results. Implement these strategies for smoother experiments and reliable data.
